9F, Zhongrui Jumei Building, 68 Jiuzhang Road, Suzhou Industrial Park, Jiangsu Province
Medical information
Chinese scientists jointly report that COVID-19 RNA does not have the ability to integrate into the host genome
SARS-CoV-2 is a highly transmissible single stranded positive stranded RNA virus that is the cause of the severe COVID-19 pandemic. SARS-CoV-2 has a highly infectious characteristic and causes severe clinical symptoms by invading multiple organ systems. This study focuses on the possibility of whether the RNA genome of COVID-19 is integrated into the genome of host cells infected by it.
On August 2, 2021, the team of Yang Yungui from the Beijing Institute of Genomics, Chinese Academy of Sciences (National Bioinformatics Center) and the team of Peng Xiaozhong from the Institute of Medical Biology, Chinese Academy of Medical Sciences worked together in Protein& Cell published online the research results entitled Comprehensive analysis of RNA-seq and whole genome sequencing data reviews no evidence for SARS-CoV-2 integrating into host genome (the work was submitted to a preprint on June 7, 2021) [1]. Through RNA sequencing (RNA-Seq) and genome wide sequencing of SARS-CoV-2 infected human and monkey cells, this work found that host virus chimeric sequences exist in the transcriptome sequencing results of SARS-CoV-2 infected cells, and in-depth research confirmed that such chimeric sequences are essentially random connecting fragments introduced during the library construction process; Through the whole genome sequencing of the host cell, it was further confirmed that there was no corresponding chimeric DNA sequence, which ruled out the possibility of integrating the SARS-CoV-2 virus RNA genome into the host DNA genome.

First, transcriptome sequencing was performed on SARS-CoV-2 infected human (293T, Huh-7, and Calu-3) and monkey cell lines (Cos-7, and MA-104). Host virus chimeric RNA fragments were identified in transcriptome sequencing results (accounting for about 0.5% to 1.8% of the total viral sequencing read segments), but these chimeric events have extremely low repeatability; Further correlation analysis of the expression level of chimeric host genes revealed that the frequency of host gene chimeric events was highly correlated with their expression level. The above results suggest that these chimeric events may not occur at the genomic DNA level and are not produced through transcription, but rather are introduced through a random pathway.
The study further analyzed the whole gene sequencing results of three human cell lines, and found no corresponding host virus chimeric genomic DNA fragments in the chromosome regions corresponding to the high-frequency chimeric genes. No DNA reads matching the SARS-CoV-2 reference genome were detected from the whole genome sequencing data. The above results indicate that SARS-CoV-2 does not have the ability to integrate into the host genome, and the chimeric sequences in transcriptome sequencing data may come from other pathways, with the greatest possibility of random linkage during the construction of RNA sequencing libraries.
In order to verify that the source of chimeric events in the transcriptome sequencing results was randomly linked and introduced by the library construction process, this study mixed the RNA from SARS-CoV-2 infected cells and uninfected zebrafish embryos, then constructed an RNA sequencing library and further sequenced the transcriptome. The results showed that in addition to the expected human virus chimeric RNA fragments, zebrafish virus chimeric RNA fragments that could not exist under natural conditions were also detected, Moreover, the number of chimeric read segments in different species is correlated with their respective RNA abundance ratios in the mixed sample, demonstrating that these chimeric read segments originate from random connections during library construction.
In summary, this study ruled out the possibility of integrating SARS-CoV-2 into the host genome through systematic analysis of transcriptome sequencing, genome wide sequencing, and transcriptome sequencing results from mixed samples (see figure below). It is noteworthy that recently, Zhou Li Quan's research team analyzed public transcriptome data and found that the expression level of retrotransposons in human cells infected with SARS-CoV-2 increased, and identified viral host chimeric RNA reading segments [2]; Rudolf Jaenisch's research team used a research system that overexpressed LINE-1 cell lines to sequence host DNA using nanopore long reading sequencing technology, identifying the 3& amp; amp; # x2032; of SARS-CoV-2 genomic RNA; rsquo; Terminal regions are integrated into the host genome through reverse transcription events [3]. Two other independent studies have found that SARS-CoV-2 is not integrated into the host cell genome, consistent with the conclusions of this study. Geoffrey J. Faulkner's research team also used nanopore long reading sequencing technology to sequence the DNA of normal HEK293T cells infected with SARS-CoV-2, and no LINE-1 mediated SARS-CoV-2 genomic integration signal was found [4]; Majid Kazemian's research team also revealed the randomness and low repeatability of host virus chimeric RNA fragments through transcriptome sequencing of SARS-CoV-2 infected cells [5].
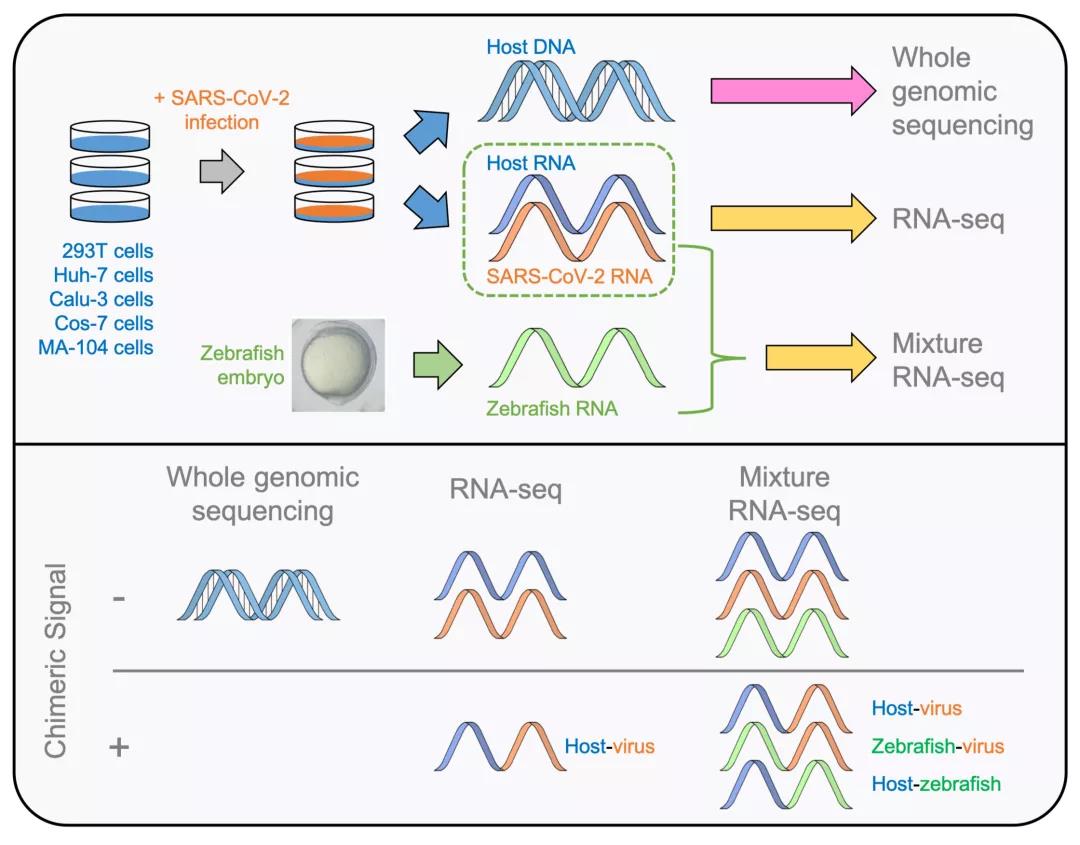
Figure: Transcriptome and whole genome sequencing revealed that novel coronavirus RNA does not have the ability to integrate into the host genome
Researcher Yang Yungui, Associate Researcher Yang Ying, and Researcher Peng Xiaozhong, Institute of Medical Biology, Chinese Academy of Sciences, Beijing Institute of Genomics (National Center for Biological Information) are the co corresponding authors of this article. Dr. Chen Yusheng, Special Research Assistant of Beijing Institute of Genomics, Chinese Academy of Sciences (National Center for Bioinformatics), Dr. Zhang Bing, Senior Engineer, and Lu Shuaiyao, Researcher of Institute of Medical Biology, Chinese Academy of Medical Sciences, are the co first author of this paper.
It is worth mentioning that a few days ago Cell Reports published online the relevant research of researchers from the University of Queensland, Australia, which also does not support the integration of COVID-19 RNA into the host genome.

